Engineering and directed evolution of protein-based tools
Proteins are the biological nano-machines that are necessary for all life on Earth. Whereas the range of functions for naturally occurring proteins is very broad, there are limits to the range observed in Nature, since proteins evolved for the sole purpose of ensuring survival of their host species. Fortunately, some natural proteins also happen to be very useful tools for research, medical, and industrial applications. Our research is focused on making them even better! By mimicking the process of evolution in the laboratory, we develop proteins that are more useful tools than their natural versions.
To date, the Campbell lab has mainly focused on engineering of fluorescent proteins. Fluorescent proteins are colorful “glowing” proteins that are found in jellyfish, sea anemone, and reef corals. These proteins have the unique ability to generate a visibly-fluorescent chromophore within their structure. The DNA that encodes a fluorescent protein can be introduced into practically any animal and be expressed using the animal’s own cellular machinery, making the tissues of the animal fluorescent. Genetically encoded fluorescence has revolutionized many areas of biology because it enables researchers to use fluorescence microscopy to “see” structures and cellular processes that would normally be invisible.
Cutting edge research in the Campbell lab involves converting fluorescent proteins into dynamic indicators that change their fluorescence intensity or color in response to a specific biochemical change. We have a particular focus on broadening the spectrum of available colors and pushing them to ever redder wavelengths. Tissue is most transparent at near-infrared wavelengths, so red-shifted indicators can let us see deeper into tissue.
The followings are highlights of our past achievements and ongoing directions.
Our current projects
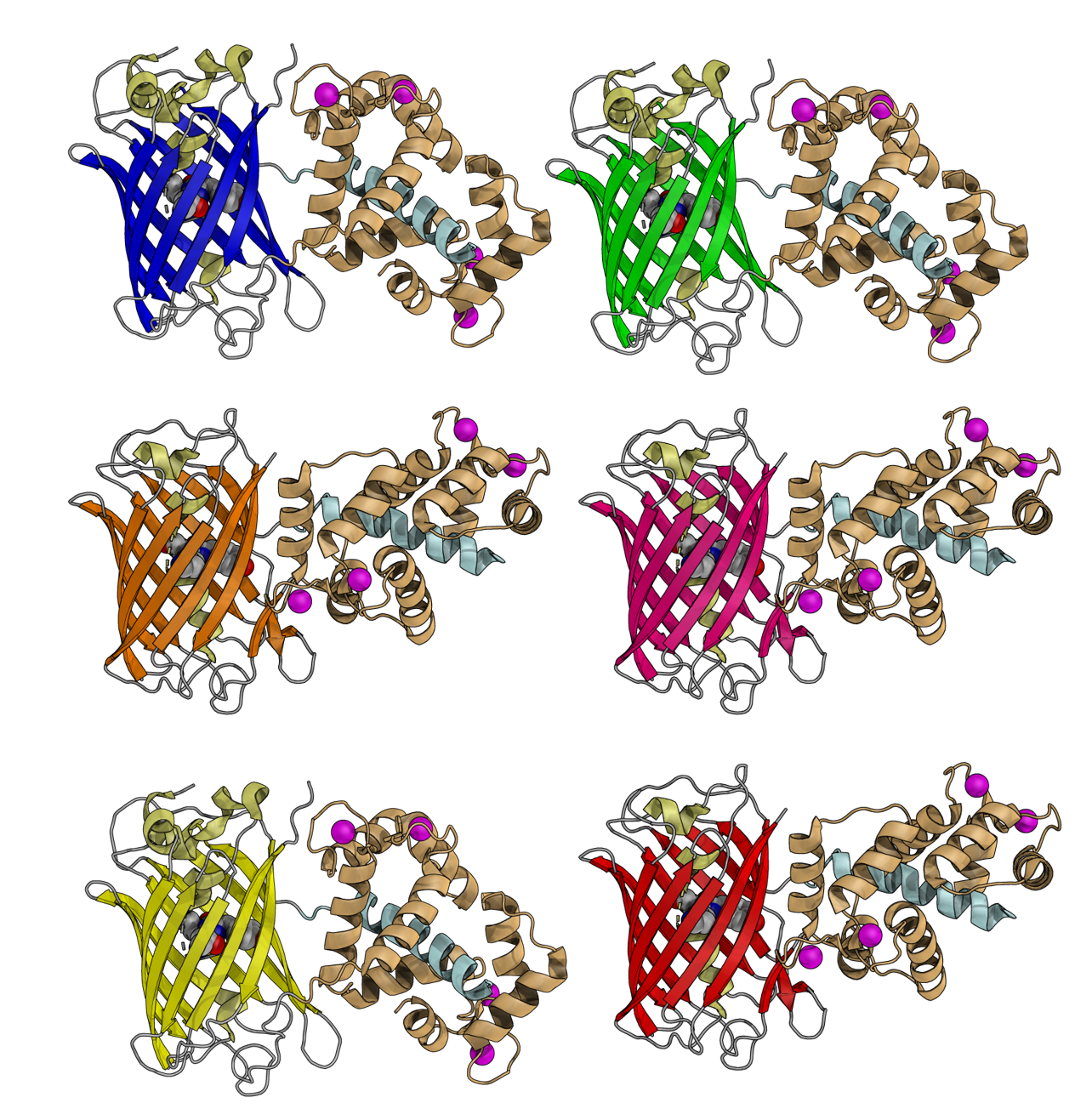
3D representations of GECO series with different colors
Engineering a palette of visible to near-infrared fluorescence neural activity indicators
Visualization of neural activity in model organisms is a powerful approach for understanding brain function and neurodegenerative diseases. To visualize neural activity, we require indicators for changes in ion concentrations, membrane voltage, or neurotransmitters. The Campbell lab launched the field of multicolor neural activity imaging with the development of a colorful palette of Ca2+ indicators known as the GECO series. We initially reported the red fluorescent R-GECO1, which has served as the basis for many other red fluorescent indicators from labs worldwide. The GECO series was further extended to be optimal for 2-photon and ratiometric imaging. Versions of R-GECO1 are freely used by hundreds of research groups around the world. Our most recent addition to the GECO series is near-infrared (NIR)-GECO1 and 2. The Campbell lab also developed several other classes of red-shifted neural activity indicators, including the NIR fluorescent QuasAr-series of voltage indicators, the red fluorescent FlicR1 voltage indicator, and a red fluorescent glutamate indicator.
Representative Publications
- Y. Zhao, et al., “An Expanded Palette of Genetically Encoded Ca2+ Indicators”, Science, 2011, 333, 1888–1891.
- J. Wu, et al., “A long Stokes shift red fluorescent protein Ca2+ indicator for 2-photon and ratiometric imaging”, Nat. Commun., 2014, 5, 5262.
- D.R. Hochbaum, et al., “All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins”, Nat. Methods, 2014, 11, 825–833.
- J. Wu, et al., “Genetically Encoded Glutamate Indicators with Altered Color and Topology”, ACS Chem. Biol.,2018, 13, 1832–1837.
- Y. Qian, et al., “A genetically encoded near-infrared fluorescent calcium ion indicator”, Nat. Methods, 2019, 16, 171–174.
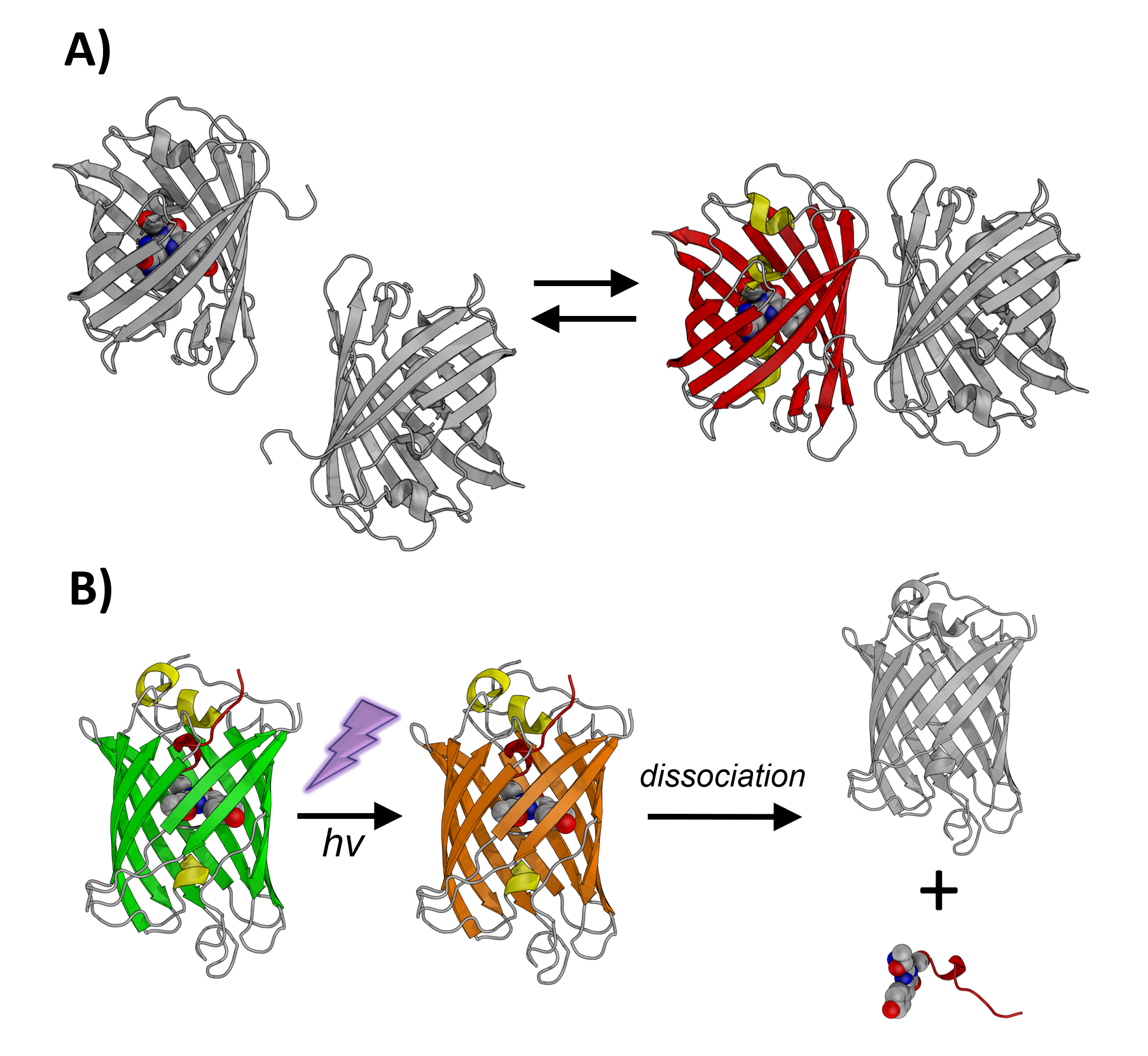
Schematic representation of A) ddFP and B) PhoCl
Pushing the limits of fluorescent protein technology using directed evolution
The Campbell lab has expanded the toolbox of coral-derived fluorescent protein variants for use in a wide variety of application areas. For example, we developed cyan, yellow, and green-to-red photoconvertible fluorescent proteins. These new fluorescent proteins enabled a number of new applications including simultaneous imaging of multiple biological processes in single cells and improved super-resolution imaging. In later work, we developed dimerization-dependent fluorescent proteins (ddFPs) that increase their brightness when they form a heterodimeric complex. This innovative technology provided a powerful new approach for detecting protein-protein interactions in live cell. Recently, the Campbell lab developed a photocleavable protein (PhoCl) as a fundamentally new class of optogenetic tool to control protein function with light. First and second generation PhoCl variants can be used for light-activated transcription, gene recombination, manipulation of protein localization, and activation of enzyme activity.
Representative Publications
- H. Hoi, et al., “A Monomeric Photoconvertible Fluorescent Protein for Imaging of Dynamic Protein Localization”, J. Mol. Biol., 2010, 401, 776–791.
- H-w. Ai, et al., “Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications”. Nat. Protocols, 2014, 9, 910–928.
- Y. Ding et al., “Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange”, Nature Methods, 2015, 12, 195–198.
- W. Zhang, et al., “Optogenetic Control with a Photocleavable Protein, PhoCl”, Nat. Methods, 2017, 14, 391-394.
- X. Lu, et al., “Photocleavable proteins that undergo fast and efficient dissociation”, Chem. Sci., 2021, 12,9658-9672.
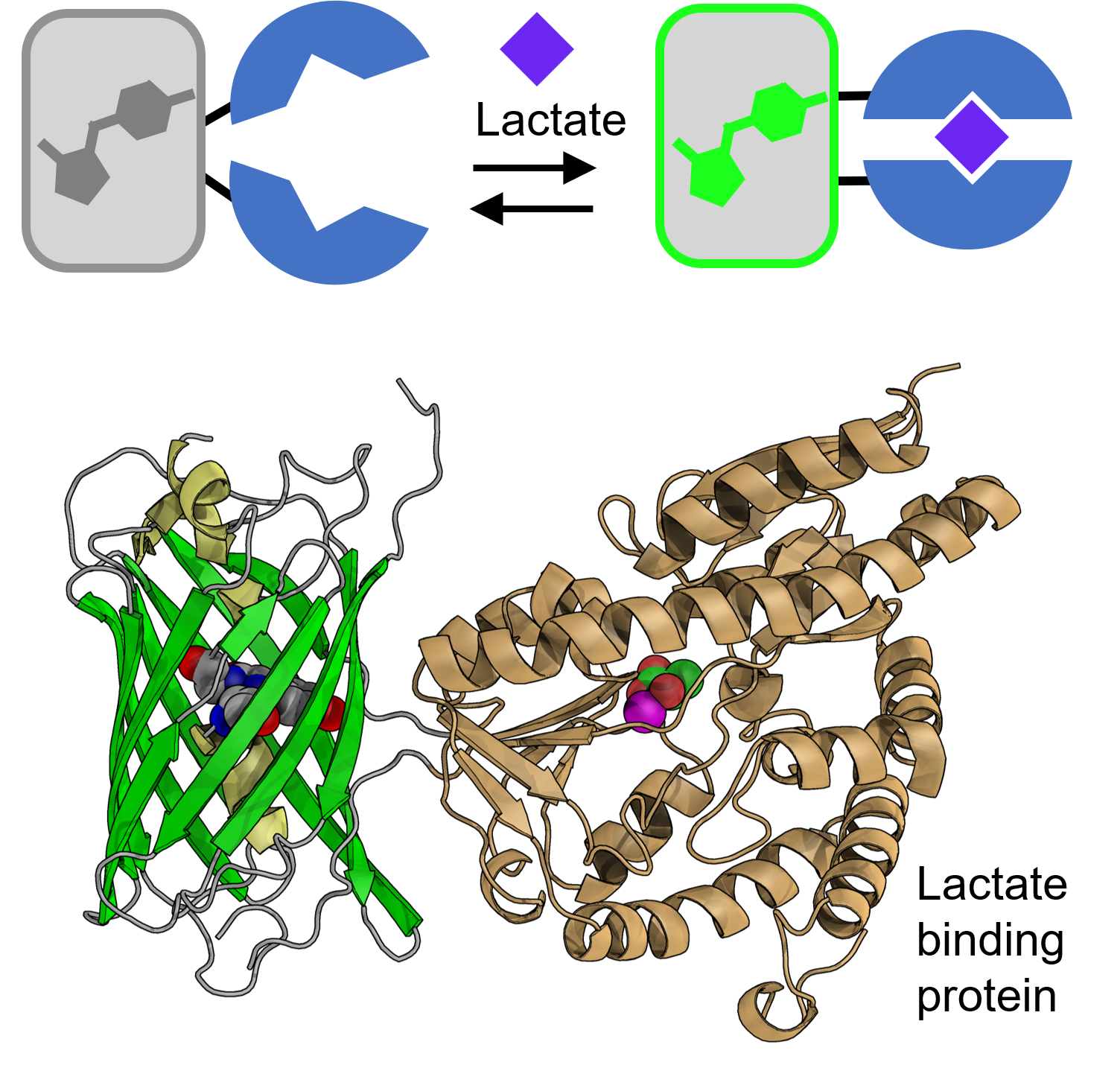
Schematic and 3D structural representations of our lactate sensor (in collaboration with Dr. M. Joanne Lemieux and Dr. Yurong Wen, University of Alberta)
Investigating the role of biological energy usage in disease
Many diseases have causes related to changes in the way cells access or use biological energy sources. One important example is neurodegenerative diseases that are associated with changes in brain energy metabolism and, specifically, the availability of lactate. Unfortunately, we currently lack tools for multiparameter visualization of the full metabolism of a cell, and so we cannot visualize the extent to which a single cell is using each of the fundamental metabolic pathways for accessing and using biological energy. The Campbell lab is developing high performance indicators for cell metabolic pathways and enable the full metabolism of a cell to be visualized for the first time!
Representative Publications
- Y. Zhao, et al., “High performance intensiometric direct- and inverse-response genetically encoded biosensors for citrate”, ACS Cent. Sci., 2020, 6, 1441–1450.
- Y. Nasu, et al., “A genetically encoded fluorescent biosensor for extracellular L-lactate“, Nat. Commun.,2021, 12, 7058.
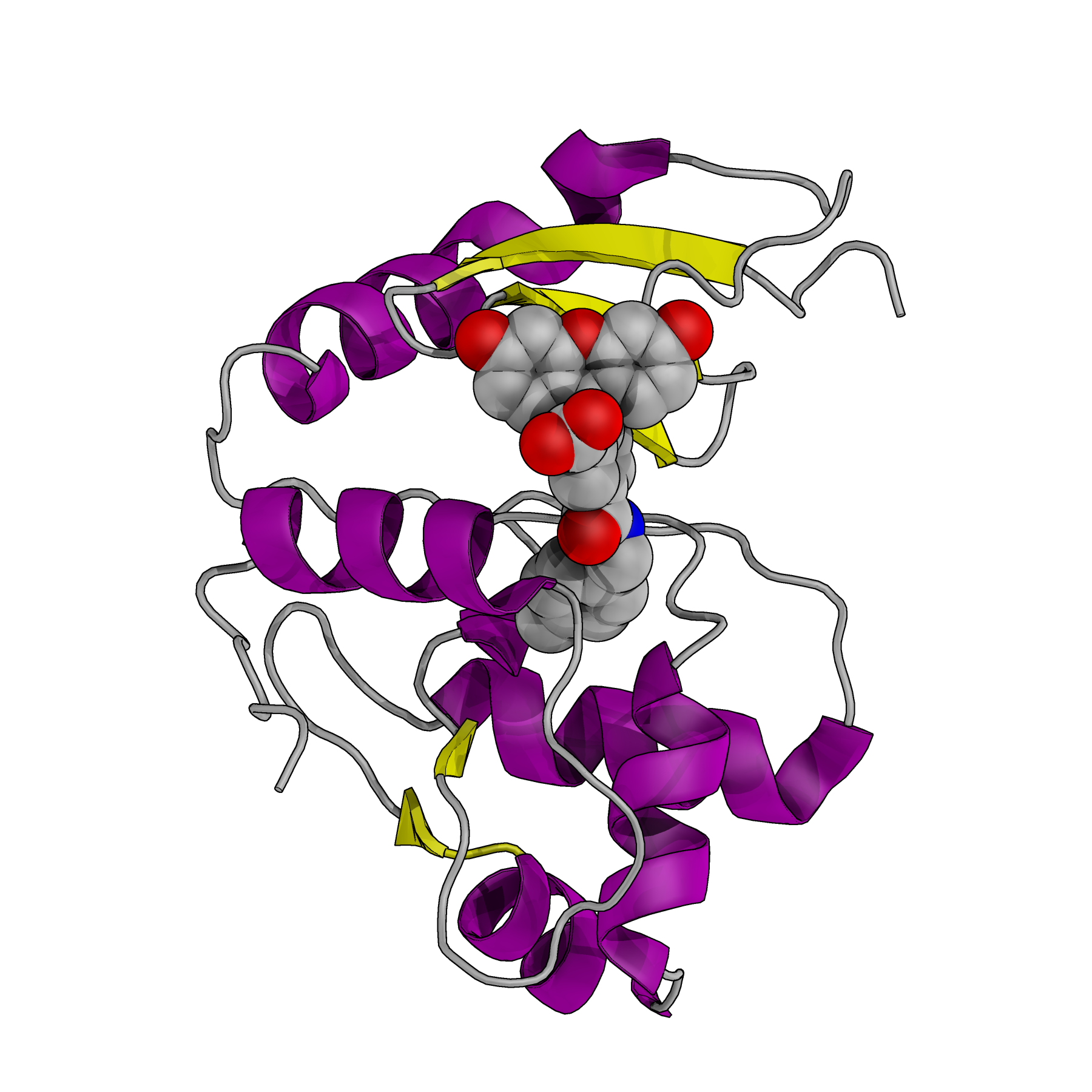
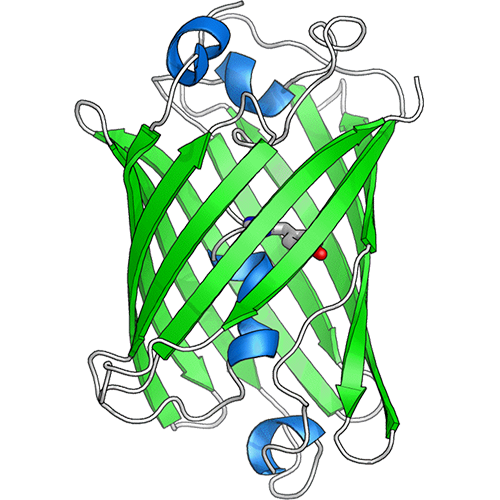
Example of chemi-genetic fluorescent protein, where a synthetic dye (space-filling model) is attached to a tag protein (PDB: 6GA0)
Indicators based on hybrid chemi-genetic fluorescent proteins
A very promising approach for expanding the range of colors for multiparameter imaging, particularly in the near-infrared (NIR) region of the spectrum, are chemi-genetic fluorescent proteins. A chemi-genetic fluorescent protein is a hybrid system in which a synthetic dye (fluorogen) is designed to attach to a specific protein expressed in the cell or tissue. Over the coming years, the Campbell lab will work to develop novel chemi-genetic fluorescent proteins and to convert them into a new generation of high-performance indicators. This is a largely unexplored and exciting research frontier.